As NGS technology has been increasingly applied in scientific research and at clinical settings, it is now possible to conduct sequencing exclusively on targeted genomic regions or a pre-defined panel of genes in large cohort of samples. As an alternative to whole genome sequencing, targeted region sequencing enables researchers to hunt for their variants of interest in pre-selected genomic regions or to validate a set of candidate hits in large cohort of samples in a cost-effective way.
Technique Highlights
– Targeted region sequencing is a prior knowledge guided approach, in contrast to the random screening approach by whole genome sequencing. For instance, the targeted regions may be chosen based on the results of previous study or may be extracted from one particular signalling pathway.
– Much less regions to be sequenced compared to exome sequencing. It significantly save research cost when applied to large quantity of samples.
– Compared to genotyping of haplotype tag SNPs, targeted region sequencing reveals the sequence of the whole gene region, thus it will not only be able to obtain the genotype of high frequency SNPs, but also has the ability to identify low frequency or sample specific unique variants.
– Compared to Sanger Sequencing based targeted sequencing, NGS based targeted sequencing is faster and more efficient when handling large number of samples.
Sample Requirement
Genomic DNA from various sources such as blood, tissue, diverse pathogenic microorganisms, and others.
Service Workflow
1. Solution hybridization based Capture array
1) Project consultation and experimental design including candidate targeted region evaluation and selection.
2) Design probes for targeted region using Aglient SureDesign Software.
3) Order Aglient SureSelect Custom targeted region capture array (through GeneSky or by customers)
4) Quality control and standardization of DNA samples.
5) gDNA Fragmentation, targeted region enrichment, library preparation. 6) Sequencing using Illumina Hiseq2000 2X100bp
7) bioinformatic analysis and filtration.
8) Genotype 12-16 high frequency SNPs to to verify NGS data quality.
2. Long PCR based enrichment system
1) Project consultation and experimental design including candidate targeted region evaluation and selection.
2) Design, synthesize and optimize long PCR primers spanning targeted region.
3) Quality control and standardization of DNA samples.
4) Long PCR amplification, PCR product fragmentation, library preparation. 5) Sequencing using Illumina Hiseq2000 2X100bp
6) bioinformatic analysis and filtration.
7) Genotype 12-16 high frequency SNPs to to verify NGS data quality.
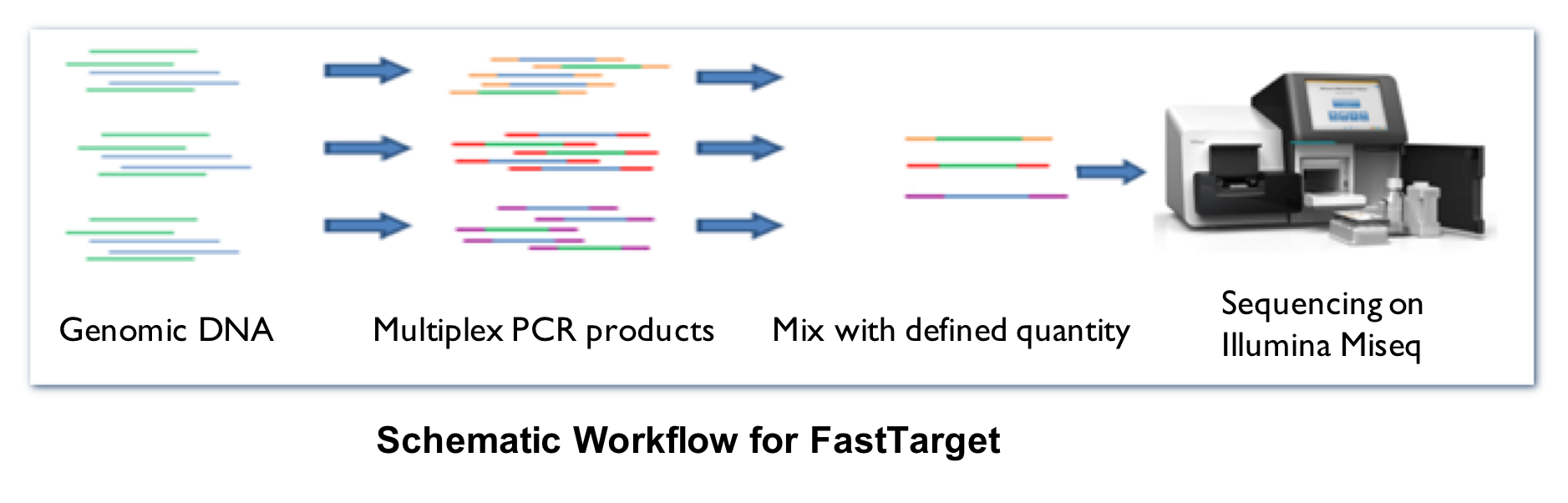
3. FastTarget enrichment system
1) Project consultation and experimental design including candidate targeted region evaluation and selection.
2) Design, synthesize and optimize multiplex-PCR primers spanning targeted region.
3) Quality control and standardization of DNA samples.
4) Multiplex-PCR amplification, sample indexing
5) Sequencing using Illumina Miseq 2X300bp
6) Bioinformatic analysis and filtration.
7) Genotype 12-16 high frequency SNPs to verify NGS data quality.
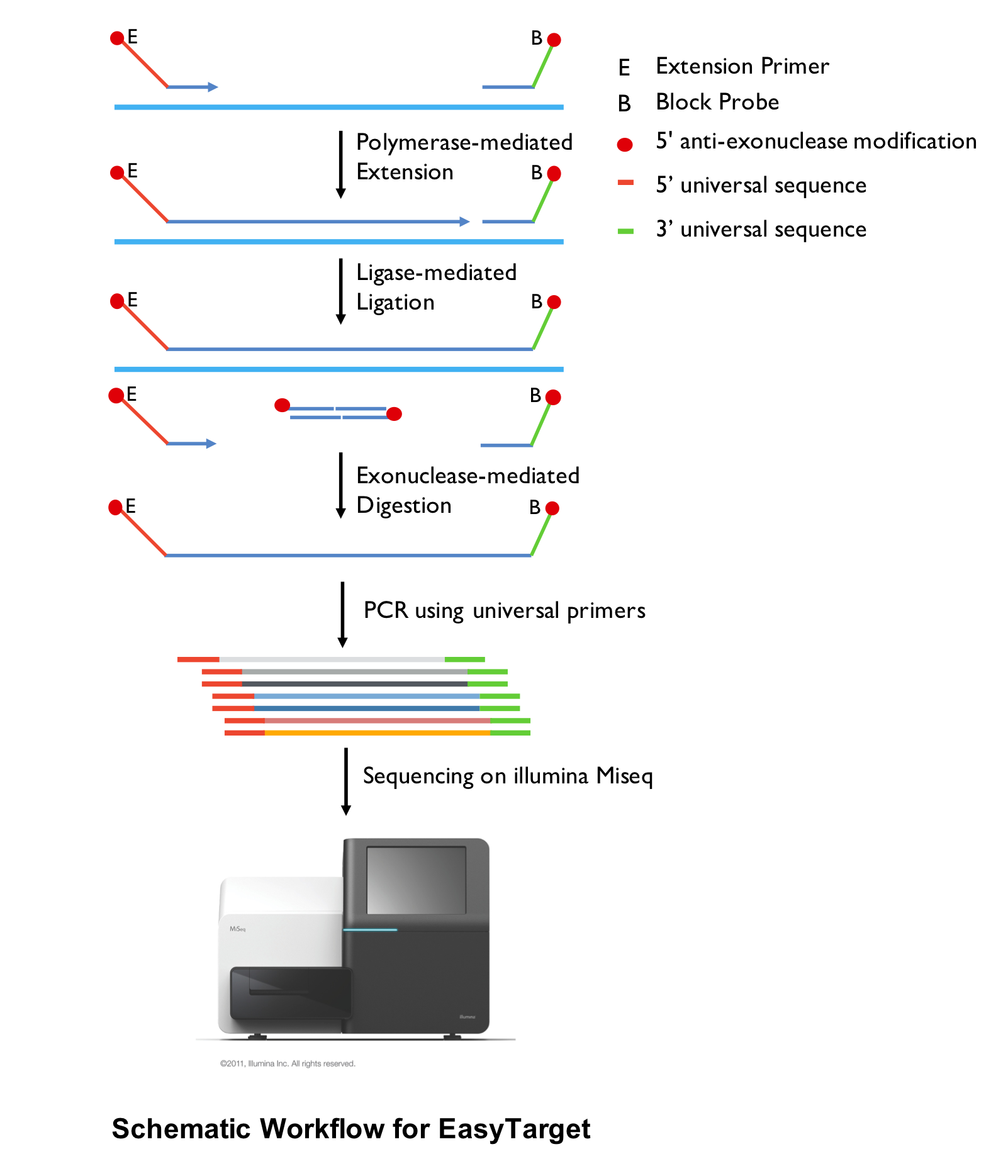
4. EasyTarget enrichment system
1) Project consultation and experimental design including candidate targeted region evaluation and selection.
2) Design, synthesize and optimize EasyTarget primers and probes spanning targeted region.
3) Quality control and standardization of DNA samples.
4) EasyTarget amplification and ligation, sample indexing.
5) Sequencing using Illumina Miseq 2X300bp.
6) Bioinformatic analysis and filtration.
7) Genotype 12-16 high frequency SNPs to to verify NGS data quality.
Deliverables
1. Experiment record
2. Quality check report