miRNA (microRNA) is a single-stranded small RNA containing 21-23 nucleotides. miRNA guides RNA- induced silencing complexes (RISC) to degrade mRNA or inhibit mRNA translation through base- pairing with target mRNA, in order to post-transcriptionally regulate protein expression. miRNA-seq based on Illumina HIseq 2000, which generates millions of miRNA sequences in each sequencing run, can repidly detect expression difference of both known and novel miRNA in different tissues, developmental stages, and pathogenic conditions. It thus provides an effective tool to investigate the role of miRNA in cellular functions and biological processes.
Technique Highlights
– High throughput Each run generates over 5 million sequencing reads.
– High resolution Recognise single nucleotide variation among miRNAs.
– High accuracy Digital counting of sequencing reads ranging from a few to hundreds of thousands. – Prior knowledge independent discovery: Identify both known and novel miRNA.
– High reproducibility Sequencing with high depth minimizes technical variations
Sample Requirement
1. Sample type: Total RNA without degradation or DNA contamination.
Service Workflow
1. Cusomtized sequencing experimental design
Library Preparation Workflow
Data Analysis Workflow
Deliverables
Standard data analysis
7. Alignment result between sequenced miRNAs and repetitive sequences, exons, introns
8. Alignment result between sequenced miRNAs and miRNAs recorded in miRBase
9. Categorization of miRNAs accord to priority
2. Starting amount of total RNA: >= 10 ug/sample
3. Sample conc.: 100 ~ 200 ng/uL
4. Sample purity: OD260/280>=1.8; 28S/18S >=1.5
5. Sample Integrity: 2100 Bioanalyzer RIN value >=8.0
2. Sample collection
3. RNA extraction and QC
4. Library preparation
5. Bioinformatics analysis
6. Project report
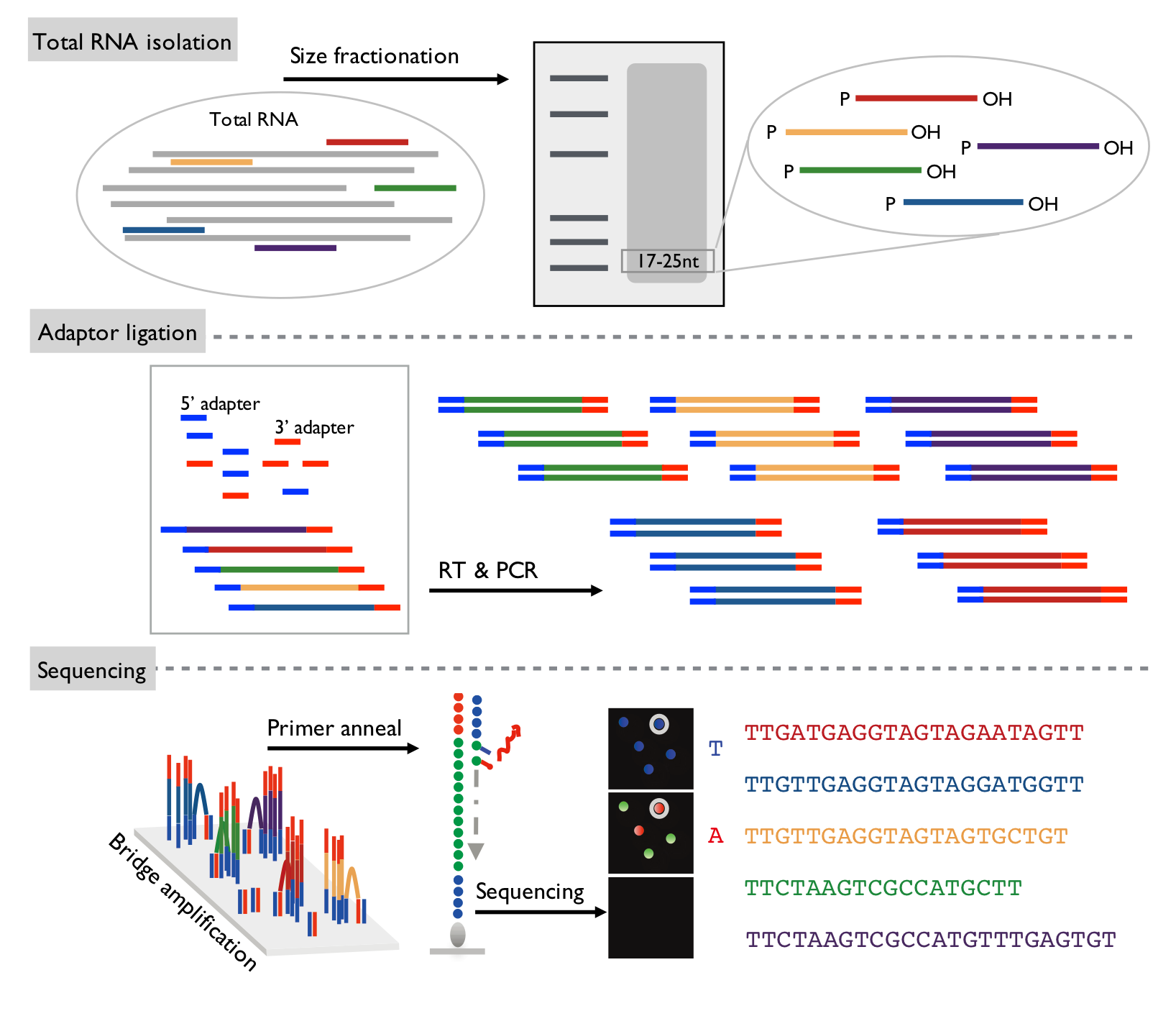
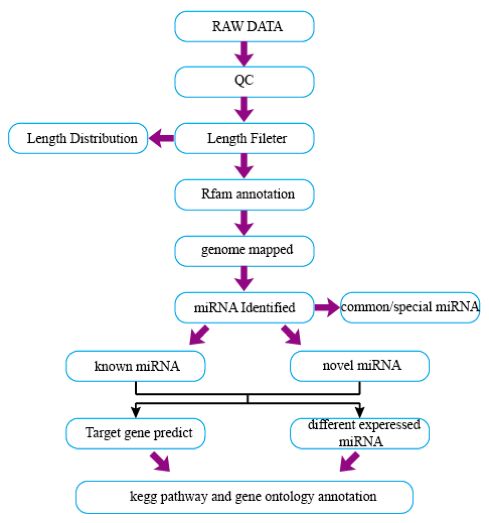
1. Raw data stored as FASTQ files
2. Overview of data output
3. 18~30nt miRNA sequencing length distribution
4. Analysis of common sequences and unique sequences between samples
5. Distribution of sequenced miRNAs that mapped to the reference genome
6. Alignment result between sequenced miRNAs and rRNAs, tRNAs, snRNAs, snoRNAs
10. Prediction of novel miRNAs from uncharacterized small RNAs using Mireap
11. Expression pattern and miRNA family analysis of known miRNA
Advanced data analysis (Based on Standard data analysis)
1. Target gene prediction of known and novel RNA
2. GO terms annotation and KEGG pathway analysis of known miRNAs
3. RNA editing analysis of known miRNAs
4. Differential expression analysis and clustering analysis of known miRNAs
5. Differential expression analysis and clustering analysis of novel miRNAs
6. GO terms annotation and KEGG pathway analysis of differentially expressed genes
Customized data analysis
We provide customized data analysis depending on specific project characteristics or upon customers’ request