
 咨询热线:400-065-6886
咨询热线:400-065-6886

 咨询热线:400-065-6886
咨询热线:400-065-6886
1.数据处理
2.概念介绍
3.PCA分析
1.不含分组信息的绘图
2.含分组信息的绘图
1.加载ggplot2包,检查输入数据和分组文件,合并数据。
2.绘制分组PCA图
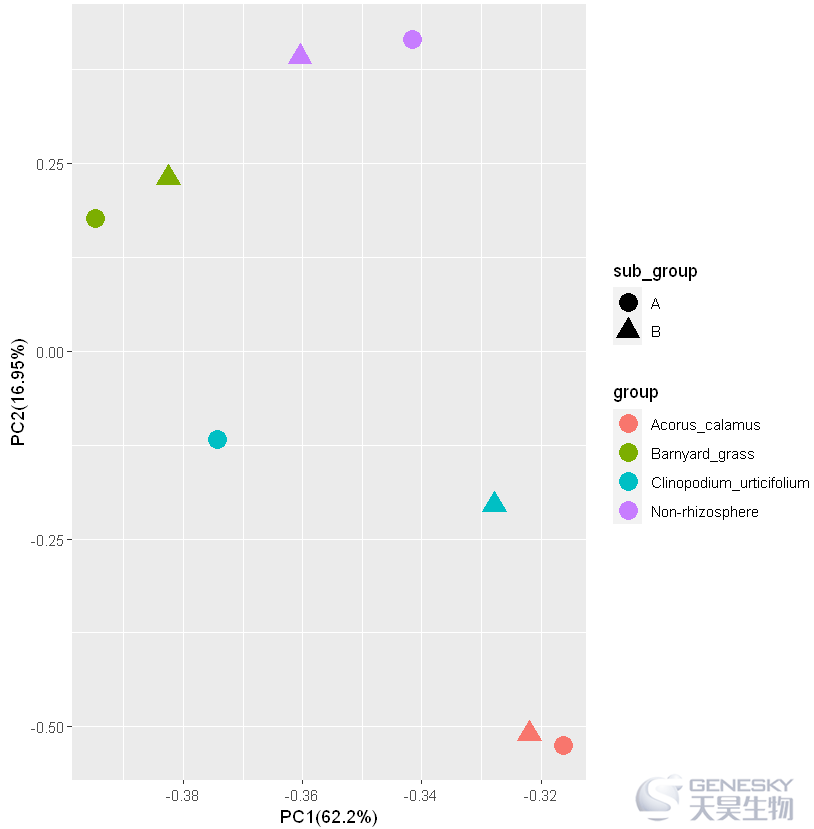
3.绘制带亚组的PCA图
OTU4 54 61 21 23 4 2 1 0 166OTU5 6 5 15 7 36 39 1 3 112OA1 A2 A3 A4 A5 A6 A7 A8OTU1 14 6 9 11 15 9 61 97OTU2 28 39 0 1 6 3 0 0OTU3 8 14 11 11 3 7 16 11OTU4 54 61 21 23 4 2 1 0OTU5 6 5 15 7 36 39 1 3OTU6 148 200 11 9 13 10 1 3data_pca = prcomp(data,scale=TRUE)
data_pcaStandard deviations (1, .., p=8):
[1] 2.2307275 1.1645475 0.9231002 0.7127234 0.3329568 0.2961777 0.2398481
[8] 0.2269067
Rotation (n x k) = (8 x 8):
PC1 PC2 PC3 PC4 PC5 PC6
A1 -0.3163354 -0.5242944 0.32746029 -0.14257138 0.04259692 -0.085790052
A2 -0.3219150 -0.5091871 0.32997621 -0.15185673 -0.03632209 0.076704375
A3 -0.3742899 -0.1172604 -0.52020283 0.06934145 0.27364994 -0.692216760
A4 -0.3277889 -0.2052396 -0.65846139 -0.04926647 -0.25746073 0.581382610
A5 -0.3947366 0.1769808 0.19555319 0.48456878 -0.08654854 0.136433532
A6 -0.3825423 0.2305448 0.15936562 0.54747900 -0.05350325 -0.003728952
A7 -0.3416118 0.4149723 0.09659485 -0.50562126 -0.61714590 -0.255016562
A8 -0.3604302 0.3915297 0.08206132 -0.39897932 0.68152299 0.293130114
PC7 PC8
A1 -0.50982176 -0.47790989
A2 0.56274135 0.42576252
A3 0.01885141 0.12780509
A4 -0.02190783 -0.09828963
A5 -0.47789491 0.53415778
A6 0.44041736 -0.52776502
A7 -0.01012305 -0.01053616
A8 0.01073030 -0.01902565| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | |
|---|---|---|---|---|---|---|---|---|
| A1 | -0.3163354 | -0.5242944 | 0.32746029 | -0.14257138 | 0.04259692 | -0.085790052 | -0.50982176 | -0.47790989 |
| A2 | -0.3219150 | -0.5091871 | 0.32997621 | -0.15185673 | -0.03632209 | 0.076704375 | 0.56274135 | 0.42576252 |
| A3 | -0.3742899 | -0.1172604 | -0.52020283 | 0.06934145 | 0.27364994 | -0.692216760 | 0.01885141 | 0.12780509 |
| A4 | -0.3277889 | -0.2052396 | -0.65846139 | -0.04926647 | -0.25746073 | 0.581382610 | -0.02190783 | -0.09828963 |
| A5 | -0.3947366 | 0.1769808 | 0.19555319 | 0.48456878 | -0.08654854 | 0.136433532 | -0.47789491 | 0.53415778 |
| A6 | -0.3825423 | 0.2305448 | 0.15936562 | 0.54747900 | -0.05350325 | -0.003728952 | 0.44041736 | -0.52776502 |
| A7 | -0.3416118 | 0.4149723 | 0.09659485 | -0.50562126 | -0.61714590 | -0.255016562 | -0.01012305 | -0.01053616 |
| A8 | -0.3604302 | 0.3915297 | 0.08206132 | -0.39897932 | 0.68152299 | 0.293130114 | 0.01073030 | -0.01902565 |
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | |
|---|---|---|---|---|---|---|---|---|
| OTU1 | -5.473904 | 3.8714451 | 1.025062 | -4.0074032 | 2.73812465 | 1.3562936 | -0.69617039 | -0.232975477 |
| OTU2 | -1.628707 | -3.1273091 | 2.262112 | -0.5625380 | -0.13332691 | 0.1758814 | 0.50210427 | 0.427617906 |
| OTU3 | -1.588757 | -0.3395338 | -0.142335 | -0.6403745 | -0.06728727 | -0.1348358 | 0.53193412 | -0.008971471 |
| OTU4 | -4.421300 | -6.2513468 | 1.470565 | -1.4061384 | -0.02607401 | -0.2792422 | 0.40394234 | 0.093156537 |
| OTU5 | -3.504735 | 1.0478361 | 0.646891 | 3.9647552 | -0.20773890 | -0.1259525 | 0.01174219 | -0.130746211 |
| OTU6 | -11.802261 | -17.2396531 | 10.654976 | -3.9452722 | -0.04030091 | 0.1222261 | 3.01654802 | 1.283238685 |
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 2.231 1.1645 0.9231 0.7127 0.33296 0.29618 0.23985
Proportion of Variance 0.622 0.1695 0.1065 0.0635 0.01386 0.01097 0.00719
Cumulative Proportion 0.622 0.7915 0.8981 0.9616 0.97541 0.98637 0.99356
PC8
Standard deviation 0.22691
Proportion of Variance 0.00644
Cumulative Proportion 1.00000
In [9]:
rotation = data_pca$rotation[,1:2]
rotation
Otu[9]:
| PC1 | PC2 | |
|---|---|---|
| A1 | -0.3163354 | -0.5242944 |
| A2 | -0.3219150 | -0.5091871 |
| A3 | -0.3742899 | -0.1172604 |
| A4 | -0.3277889 | -0.2052396 |
| A5 | -0.3947366 | 0.1769808 |
| A6 | -0.3825423 | 0.2305448 |
| A7 | -0.3416118 | 0.4149723 |
| A8 | -0.3604302 | 0.3915297 |
1.不含分组信息的绘图
In [10]:
plot(rotation)
Otu[10]:
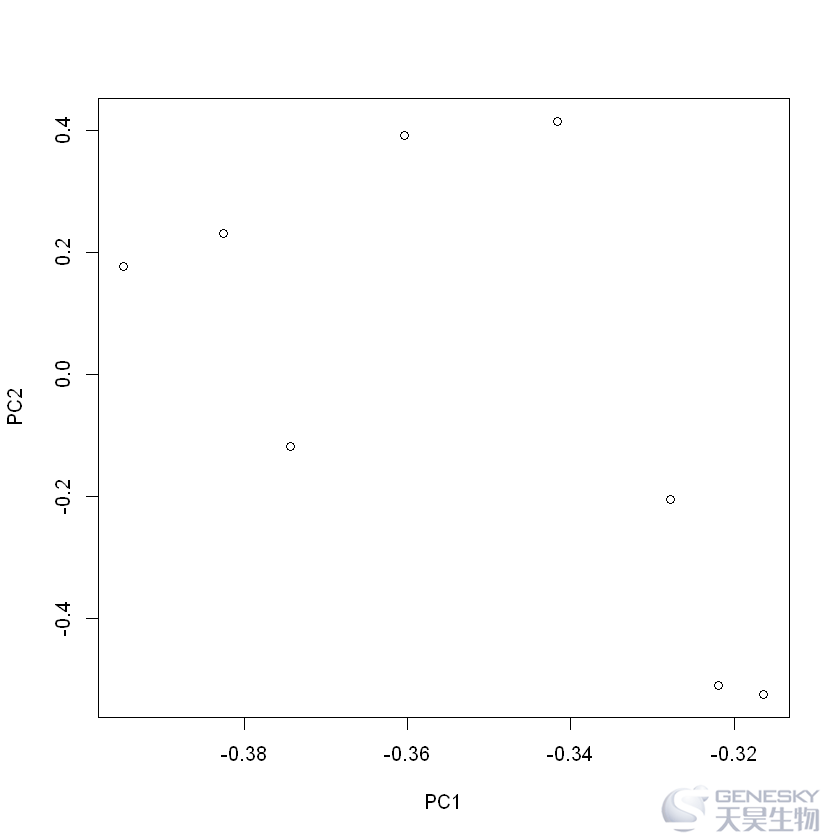
2. 含分组信息的绘图
In [11]:
group = read.table('sample.groups.xls',header = FALSE)
colnames(group)=c('sample','group','sub_group')
group
Otu[11]:
| sample | group | sub_group |
|---|---|---|
| A1 | Acorus_calamus | A |
| A2 | Acorus_calamus | B |
| A3 | Clinopodium_urticifolium | A |
| A4 | Clinopodium_urticifolium | B |
| A5 | Barnyard_grass | A |
| A6 | Barnyard_grass | B |
| A7 | Non-rhizosphere | A |
| A8 | Non-rhizosphere | B |
In [12]:
pcolor_group = rainbow(length(unique(group$group))) #利用rainbow函数选择颜色
pcolor_group
Otu[12]:
'#FF0000FF' '#80FF00FF' '#00FFFFFF' '#8000FFFF'
In [13]:
color = color_group[as.numeric(factor(group$group))]color
Otu[13]:
'#FF0000FF' '#FF0000FF' '#00FFFFFF' '#00FFFFFF' '#80FF00FF' '#80FF00FF' '#8000FFFF' '#8000FFFF'
In [14]:
plot(rotation,col = color ,pch = c(21,22,23,24)[group$group])
legend('topright',legend = levels(group$group),col = color_group,pch=c(21,22,23,24))
title('PCA')
Otu[14]:
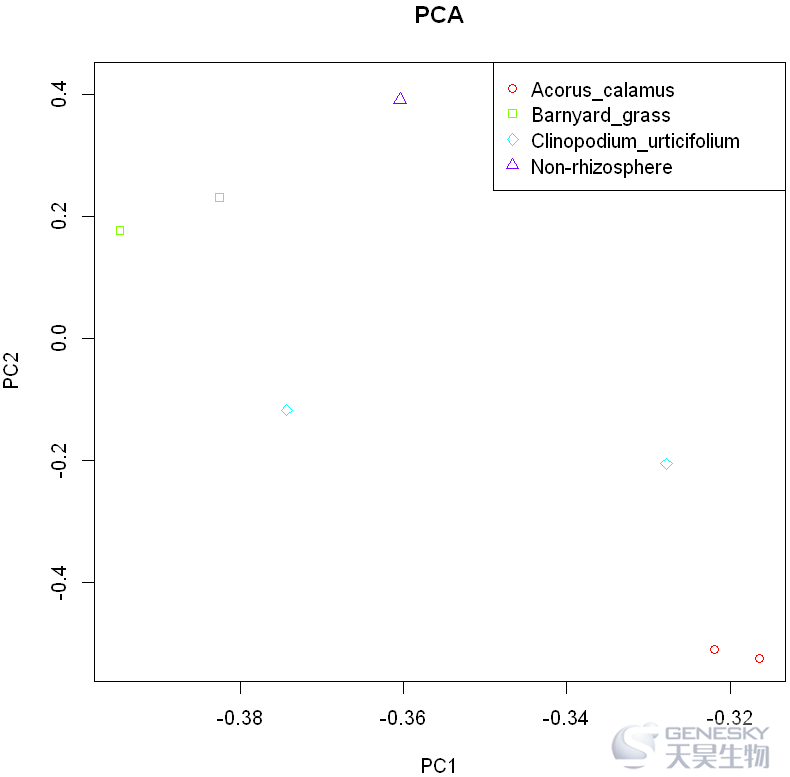
1. 加载ggplot2包,检查输入数据和分组文件,合并数据。
In [15]:
install.packages('ggplot2') #未安装这个包的话,需要先安装ggplot2
library(ggplot2)
Warning message:
"package 'ggplot2' was built under R version 3.6.3"
| PC1 | PC2 | |
|---|---|---|
| A1 | -0.3163354 | -0.5242944 |
| A2 | -0.3219150 | -0.5091871 |
| A3 | -0.3742899 | -0.1172604 |
| A4 | -0.3277889 | -0.2052396 |
| A5 | -0.3947366 | 0.1769808 |
| A6 | -0.3825423 | 0.2305448 |
| A7 | -0.3416118 | 0.4149723 |
| A8 | -0.3604302 | 0.3915297 |
Group =read.table('sample.groups.xls',header = FALSE)colnames(Group) = c('sample','group','sub_group')Group| sample | group | sub_group |
|---|---|---|
| A1 | Acorus_calamus | A |
| A2 | Acorus_calamus | B |
| A3 | Clinopodium_urticifolium | A |
| A4 | Clinopodium_urticifolium | B |
| A5 | Barnyard_grass | A |
| A6 | Barnyard_grass | B |
| A7 | Non-rhizosphere | A |
| A8 | Non-rhizosphere | B |
pca_result = cbind(rotation,Group)pca_result = data.frame(pca_result)pca_result| PC1 | PC2 | sample | group | sub_group | |
|---|---|---|---|---|---|
| A1 | -0.3163354 | -0.5242944 | A1 | Acorus_calamus | A |
| A2 | -0.3219150 | -0.5091871 | A2 | Acorus_calamus | B |
| A3 | -0.3742899 | -0.1172604 | A3 | Clinopodium_urticifolium | A |
| A4 | -0.3277889 | -0.2052396 | A4 | Clinopodium_urticifolium | B |
| A5 | -0.3947366 | 0.1769808 | A5 | Barnyard_grass | A |
| A6 | -0.3825423 | 0.2305448 | A6 | Barnyard_grass | B |
| A7 | -0.3416118 | 0.4149723 | A7 | Non-rhizosphere | A |
| A8 | -0.3604302 | 0.3915297 | A8 | Non-rhizosphere |
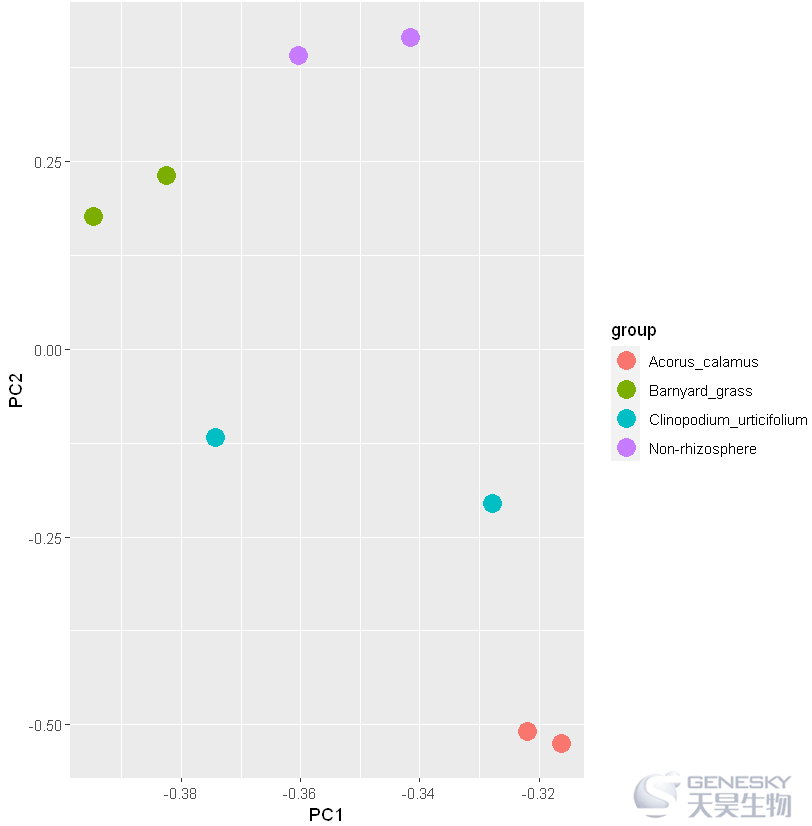
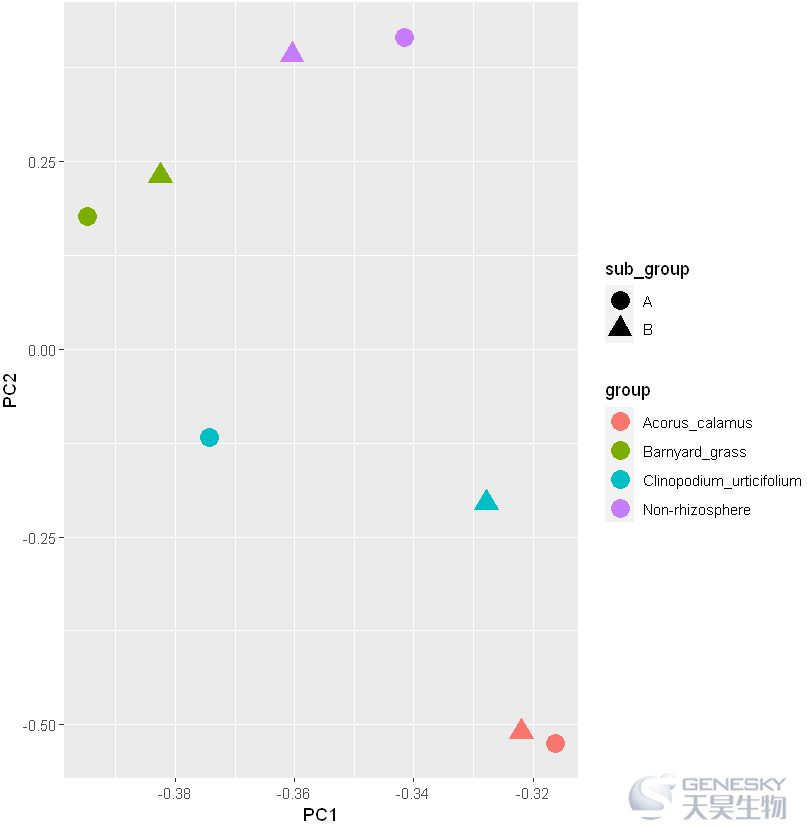
往期链接:
如果您对本文案介绍的方法或代码有疑问,
请扫码添加QQ群沟通
【本群将为大家提供】
分享生信分析方案
提供数据素材及分析软件支持
定期开展生信分析线上讲座

QQ号:1040471849
作者:大熊
审核:有才
来源:天昊生信团
微信扫一扫
关注该公众号
前往“发现”-“看一看”浏览“朋友在看”