咨询热线:400-065-6886
咨询热线:400-065-6886

 咨询热线:400-065-6886
咨询热线:400-065-6886
文章导读
2. 调整短轴和长轴的参数
一
数据处理
1.otu文件、分组文件查看
In [1]:
df = read.table('otu.tax.0.03.xls',header = T,check.names = F,sep = ' ',
row.names = 1,quote = '',comment.char = '')
head(df,3)
Out[1]:
B1 B10 B11 B12 B13 B14 B16 B18 B19 B2 ... F9 OTUsize Taxonomy superkingdom phylum class order family genus species
OTU1 2004 21 11 0 1 3 195 2 1 0 ... 0 195460 Bacteria{superkingdom}(100);Firmicutes{phylum}(100);Bacilli{class}(100);Lactobacillales{order}(100);Streptococcaceae{family}(100);Streptococcus{genus}(100);Streptococcus_anginosus{species}(100) Bacteria Firmicutes Bacilli Lactobacillales Streptococcaceae Streptococcus Streptococcus_anginosus
OTU2 5192 0 10 0 4 0 20 0 0 0 ... 0 5844 Bacteria{superkingdom}(100);Actinobacteria{phylum}(100);Actinobacteria{class}(100);Corynebacteriales{order}(100);Corynebacteriaceae{family}(100);Corynebacterium{genus}(100);Corynebacterium_amycolatum{species}(83) Bacteria Actinobacteria Actinobacteria Corynebacteriales Corynebacteriaceae Corynebacterium Corynebacterium_amycolatum
OTU3 61114 66 5850 113253 2 2 207 36 121 0 ... 0 804859 Bacteria{superkingdom}(100);Actinobacteria{phylum}(100);Actinobacteria{class}(100);Bifidobacteriales{order}(100);Bifidobacteriaceae{family}(100);Gardnerella{genus}(100);Gardnerella_vaginalis{species}(100) Bacteria Actinobacteria Actinobacteria Bifidobacteriales Bifidobacteriaceae Gardnerella Gardnerella_vaginalis
In [2]:
info = read.table('sample_group.txt')
head(info,5)
Out[2]:
V1 V2
B1 BB
10 BB
11 BB
12 BB
13 B
In [3]:
group=info$V2
levels(group)
Out[3]:
'B' 'C' 'D' 'E' 'F' 'G'
2.从OTU文件中将属于同一组的样品数据提出,并求和
In [4]:
info[info$V2=='B','V1']
Out[4]:
B1 B10 B11 B12 B13 B14 B16 B18 B19 B2 B22 B23 B26 B27 B28 B3 B4 B41 B45 B46 B49 B5 B50 B51 B6 B7 B8 B9
In [5]:
head(df[,info[info$V2=='B','V1']],5)
Out[5]:
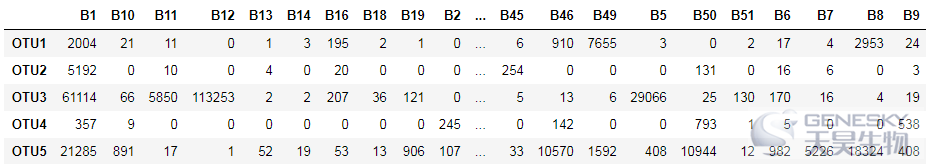
In [6]:
df$B = apply(df[,info[info$V2=='B','V1']],1,sum)
df$C = apply(df[,info[info$V2=='C','V1']],1,sum)
df$D = apply(df[,info[info$V2=='D','V1']],1,sum)
df$E = apply(df[,info[info$V2=='E','V1']],1,sum)
df$F = apply(df[,info[info$V2=='F','V1']],1,sum)
df$G = apply(df[,info[info$V2=='G','V1']],1,sum)
In [7]:
head(df,3)
Out[7]:
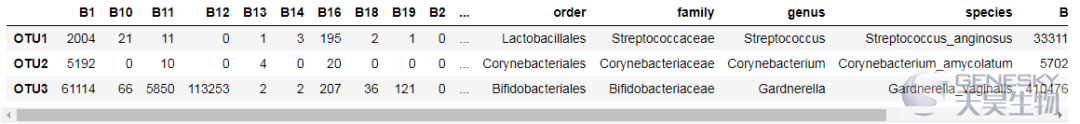
3.统计不同分组的OTU个数
In [8]:
length(rownames(df)[df$B!=0])
Out[8]:
1370
In [9]:
length(rownames(df)[df$C!=0])
Out[9]:
894
In [10]:
length(rownames(df)[df$D!=0])
Out[10]:
1630
In [11]:
length(rownames(df)[df$E!=0])
Out[11]:
855
In [12]:
length(rownames(df)[df$F!=0])
Out[12]:
684
In [13]:
length(rownames(df)[df$G!=0])
Out[13]:
473
二
venn图绘制
1.安装和加载VennDiagram
In [14]:
install.packages('VennDiagram')
In [15]:
library(VennDiagram)
2.绘图
In [16]:
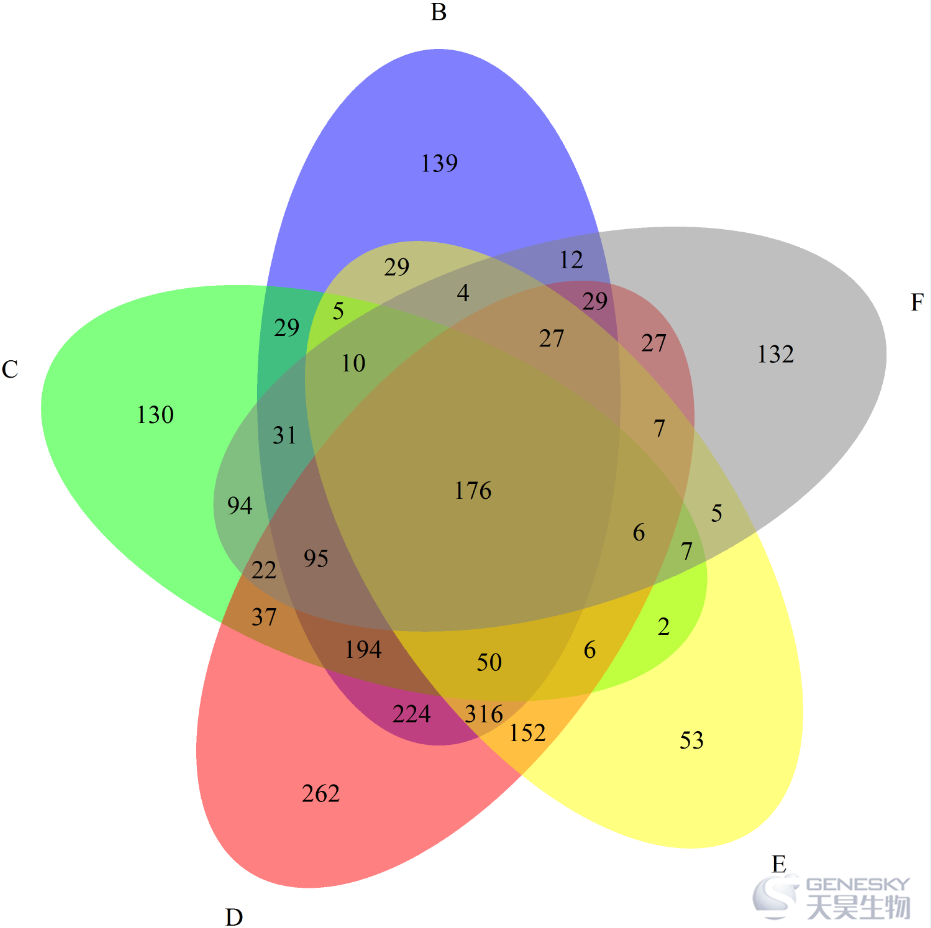
x= list(B=rownames(df)[df$B!=0],
C=rownames(df)[df$C!=0],
D=rownames(df)[df$D!=0],
E=rownames(df)[df$E!=0],
F=rownames(df)[df$F!=0])
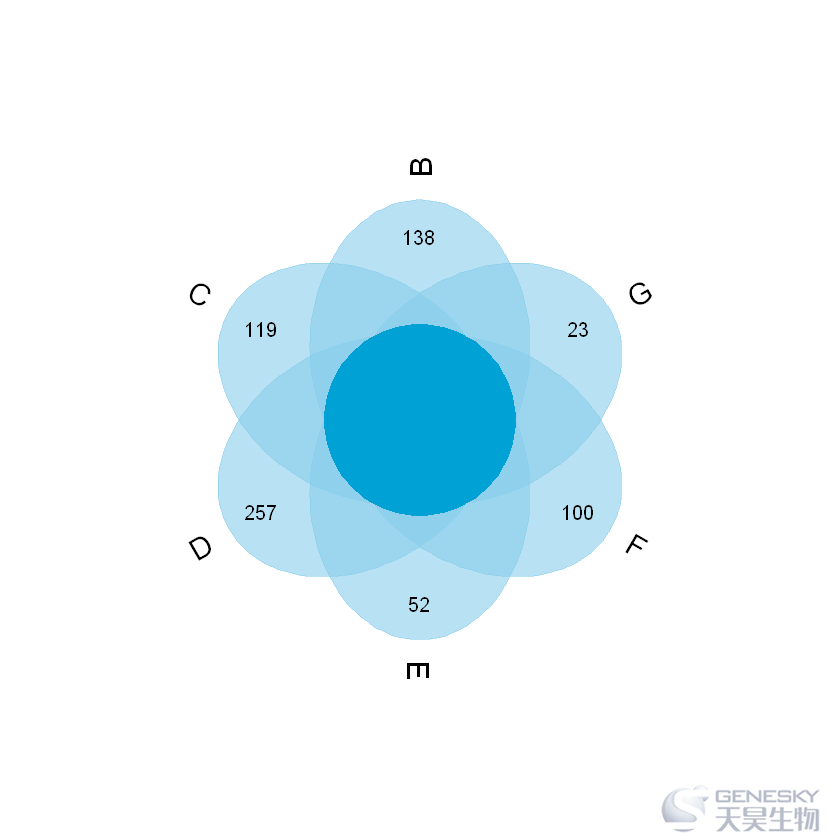
venn.diagram(x,filename = 'venn_5set.png',col='transparent',fill=c('blue','green','red','yellow','grey50'))
Out[16]:
x= list(B=rownames(df)[df$B!=0],
C=rownames(df)[df$C!=0],
D=rownames(df)[df$D!=0],
E=rownames(df)[df$E!=0])venn.diagram(x,filename = 'venn_4set.png',col='transparent',fill=c('blue','green','red','yellow'))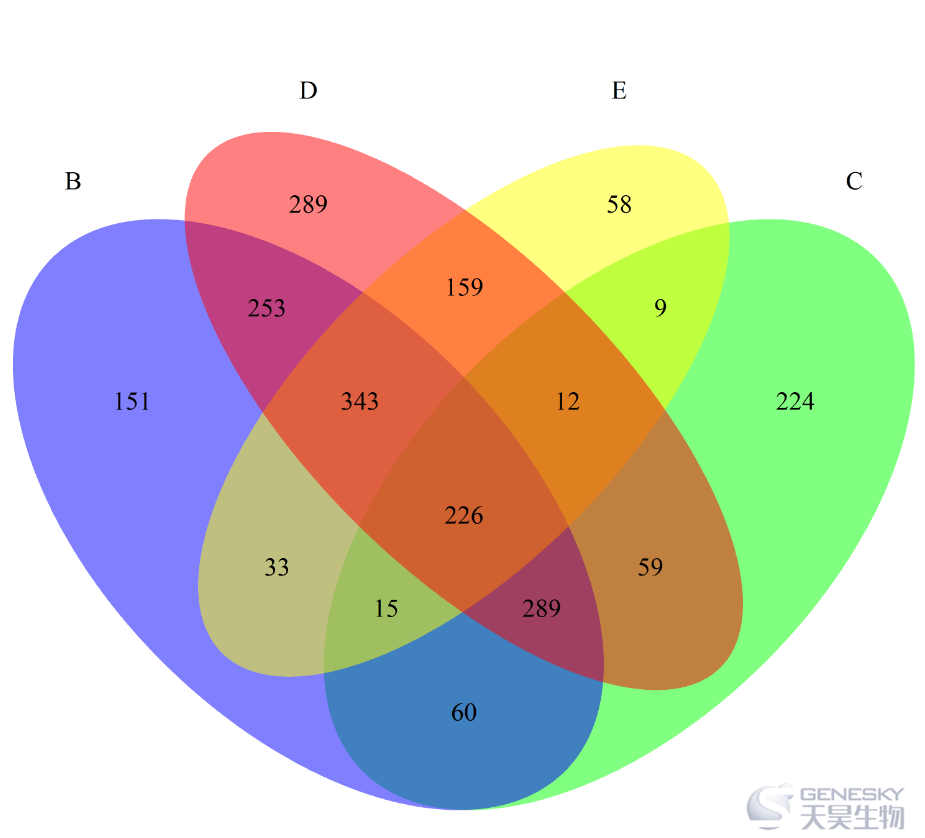
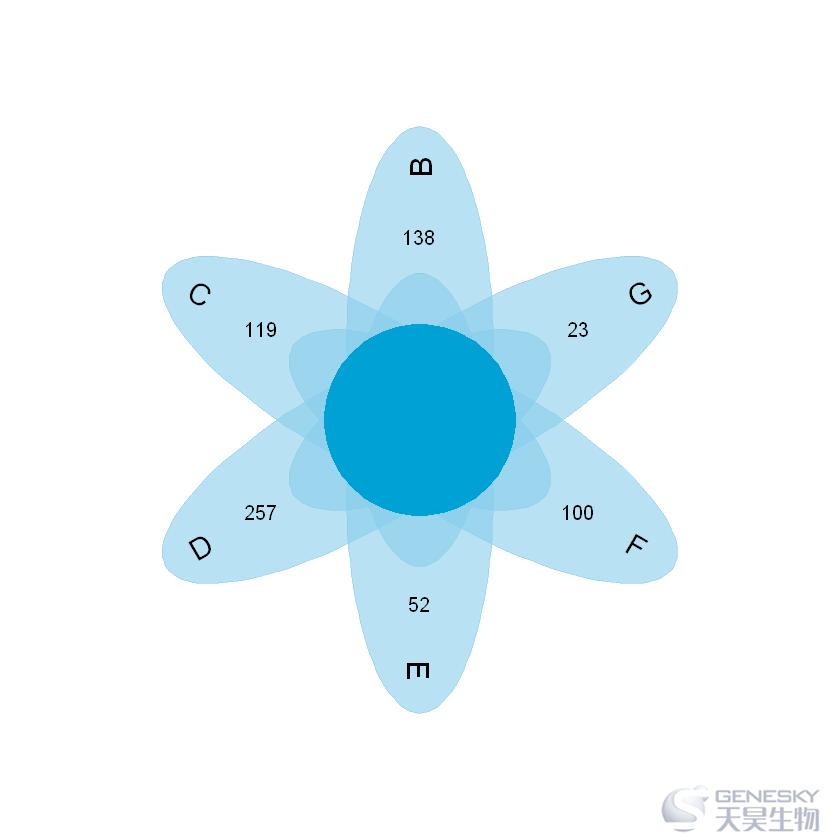
length(rownames(df)[df$B!=0 & df$C==0 & df$D!=0 & df$E==0 & df$F==0])install.packages('plotrix')library(plotrix)flower_plot <- function(sample, value, start, a, b,
ellipse_col = rgb(135, 206, 235, 150, max = 255),
circle_col = rgb(0, 162, 214, max = 255),
circle_text_cex = 1.5
{
bty = "n", ann = F, xaxt = "n", yaxt = "n", mar = c(1,1,1,1))
="n")
n <- length(sample)
deg <- 360 / n
res <- lapply(1:n, function(t){
= 5 + cos((start + deg * (t - 1)) * pi / 180),
y = 5 + sin((start + deg * (t - 1)) * pi / 180),
col = ellipse_col,
border = ellipse_col,
a = a, b = b, angle = deg * (t - 1))
= 5 + 2.5 * cos((start + deg * (t - 1)) * pi / 180),
y = 5 + 2.5 * sin((start + deg * (t - 1)) * pi / 180),
value[t]
)
if (deg * (t - 1) < 180 && deg * (t - 1) > 0 ) {
= 5 + 3.3 * cos((start + deg * (t - 1)) * pi / 180),
y = 5 + 3.3 * sin((start + deg * (t - 1)) * pi / 180),
sample[t],
srt = deg * (t - 1) - start,
adj = 1,
cex = circle_text_cex
)
else {
= 5 + 3.3 * cos((start + deg * (t - 1)) * pi / 180),
y = 5 + 3.3 * sin((start + deg * (t - 1)) * pi / 180),
sample[t],
srt = deg * (t - 1) + start,
adj = 0,
cex = circle_text_cex
)
})
= 5, y = 5, r = 1.3, col = circle_col, border = circle_col)
}b=length(rownames(df)[df$B!=0 & df$C==0 & df$D==0 & df$E==0 & df$F==0 & df$G==0 ])c=length(rownames(df)[df$B==0 & df$C!=0 & df$D==0 & df$E==0 & df$F==0 & df$G==0 ])d=length(rownames(df)[df$B==0 & df$C==0 & df$D!=0 & df$E==0 & df$F==0 & df$G==0 ])e=length(rownames(df)[df$B==0 & df$C==0 & df$D==0 & df$E!=0 & df$F==0 & df$G==0 ])f=length(rownames(df)[df$B==0 & df$C==0 & df$D==0 & df$E==0 & df$F!=0 & df$G==0 ])g=length(rownames(df)[df$B==0 & df$C==0 & df$D==0 & df$E==0 & df$F==0 & df$G!=0 ])