
 咨询热线:400-065-6886
咨询热线:400-065-6886

 咨询热线:400-065-6886
咨询热线:400-065-6886
常规的biplot绘图,虽然可以把样品和物种标在PCA图上,但是效果非常不理想。ggbiplot是一款PCA分析结果可视化的R包工具,可以直接采用ggplot2来可视化R中基础函数prcomp()结果,按分组绘图,添加椭圆、箭头和物种名称。
文章导读

1.数据读取
In [1]:
df = read.table('phylum_taxon_abundance.xls',header = T,row.names = 1)
head(df)
Out[1]:
X68 X69 X106 X6 X74 X112 X75 X77 X76 X7 X104 X105 X107 X80 X110 X111
Proteobacteria 126573 92545 83038 75739 103697 116154 73868 76555 47648 61165 90984 86653 111958 95141 92972 102300
Actinobacteria 12759 10186 50860 32437 34212 21024 64078 52398 46109 38587 46147 46310 46780 50249 32803 29345
Bacteroidetes 18428 25557 16559 22848 7046 23059 4974 8066 4984 15941 14064 12966 10931 5047 14100 27364
Acidobacteria 16566 26529 6636 16079 7563 10637 8352 11404 11755 7008 8292 9372 6405 5773 10853 6738
Chloroflexi 6665 11038 12520 20040 22476 4146 23314 26628 32961 30103 14238 11799 9819 15672 7421 6116
Firmicutes 6033 13830 21344 3293 18031 10253 12822 5480 35773 6208 13005 20341 7953 21295 19200 12794
2.PCA分析
In [2]:
data_pca = prcomp(df,scale=TRUE)
In [3]:
data_pca$rotation
Out[3]:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10 PC11 PC12 PC13 PC14 PC15 PC16
X68 -0.2430500 0.38916585 0.023282518 0.05234324 0.36699759 0.05506225 -0.050560004 -0.10593654 -0.11444462 0.36953735 -0.30537404 0.338903555 0.41402670 -0.284624230 -0.090105023 -0.12800184
X69 -0.2402164 0.36218483 -0.013061765 0.51194696 -0.07174752 0.45384168 0.176893288 -0.18258457 0.11230165 0.10908458 0.25096766 -0.373622075 -0.11261695 0.091127424 0.058957993 -0.14682460
X106 -0.2557063 -0.12447526 0.173438926 -0.13426542 -0.44009941 -0.03620825 -0.171205288 -0.27565035 -0.07107470 0.06744872 0.24642356 -0.132560788 0.22757381 -0.559334120 0.272614351 0.20942325
X6 -0.2536670 0.05623670 -0.468005262 0.15240255 -0.21849821 0.21291012 -0.042266340 0.08882465 -0.05819861 -0.63923630 -0.04586830 0.368626963 0.18738234 -0.041111689 0.002259739 0.02606331
X74 -0.2566932 0.04852627 0.119204747 0.06838263 0.54204129 -0.26228748 -0.168889220 -0.15750400 0.37401913 -0.24653397 0.43918652 0.132795250 -0.06540488 0.027949070 0.035667605 0.28180399
X112 -0.2484910 0.32559769 0.059756361 -0.02354247 -0.01974062 -0.15863691 -0.236439490 0.13963796 -0.52135144 -0.02553914 -0.12506557 -0.257332430 0.01738885 0.385639179 0.082978501 0.46575660
X75 -0.2450634 -0.34701579 -0.056762940 -0.29495653 0.02895928 0.34410897 0.015339684 0.05938054 0.25976962 0.18702376 0.06575778 -0.115475427 0.53309938 0.434943704 -0.057135102 0.09767001
X77 -0.2513740 -0.22241477 -0.275059754 -0.16383243 0.24778626 0.37816523 -0.304629744 0.31352885 -0.15620294 0.21428547 0.08568085 -0.041715898 -0.46765947 -0.285282621 -0.088286154 0.04530152
X76 -0.2184556 -0.53765837 0.359586523 0.60243062 0.04023701 -0.03870977 -0.169555479 0.06393069 -0.03699587 -0.05759508 -0.35819968 -0.007589647 0.01859631 0.003219111 -0.037389048 -0.01473870
X7 -0.2449620 -0.21491066 -0.616404879 0.17990580 0.01817630 -0.53453347 0.281215337 -0.09008429 -0.05890281 0.27059539 0.03529958 -0.167312985 0.02918828 -0.001724188 0.002416606 -0.03795324
X104 -0.2587459 -0.03355179 0.008686834 -0.16896009 -0.08510343 0.03558645 -0.005192017 -0.29396775 0.09086539 0.13010662 -0.24205025 0.337891028 -0.35686557 0.297527487 0.611915023 -0.14174029
X105 -0.2577352 -0.06682350 0.184230342 -0.10012624 -0.21442142 0.04157705 0.311069102 -0.32783475 -0.16500884 0.07210566 0.08267707 0.313885377 -0.25793701 0.103284263 -0.617888919 0.19085013
X107 -0.2568870 0.06880793 0.019220849 -0.32495584 0.14486672 -0.01338294 0.153501873 -0.19078478 0.22179854 -0.35954343 -0.50832904 -0.485621957 -0.10601452 -0.191133770 -0.135823579 -0.05717753
X80 -0.2564216 -0.09961860 0.240161326 -0.18274753 0.16787236 -0.09335768 0.143164978 0.09703855 -0.45169269 -0.22824042 0.32685081 -0.057313696 0.09276323 0.064916502 0.064533540 -0.61904805
X110 -0.2565415 0.11833528 0.226514035 -0.01270806 -0.10483179 -0.07664522 0.524717549 0.65239299 0.21146831 0.06159903 -0.01427284 0.115320466 -0.04797013 -0.149323315 0.172812582 0.18554028
X111 -0.2528704 0.22570474 0.029792623 -0.04277772 -0.38337351 -0.28980173 -0.483304622 0.21887859 0.35215399 0.11053195 0.01820724
0.002325398 -0.06549265 0.117608596 -0.291131534 -0.36124728
3.biplot图绘制
In [4]:
biplot(data_pca)
Out[4]:
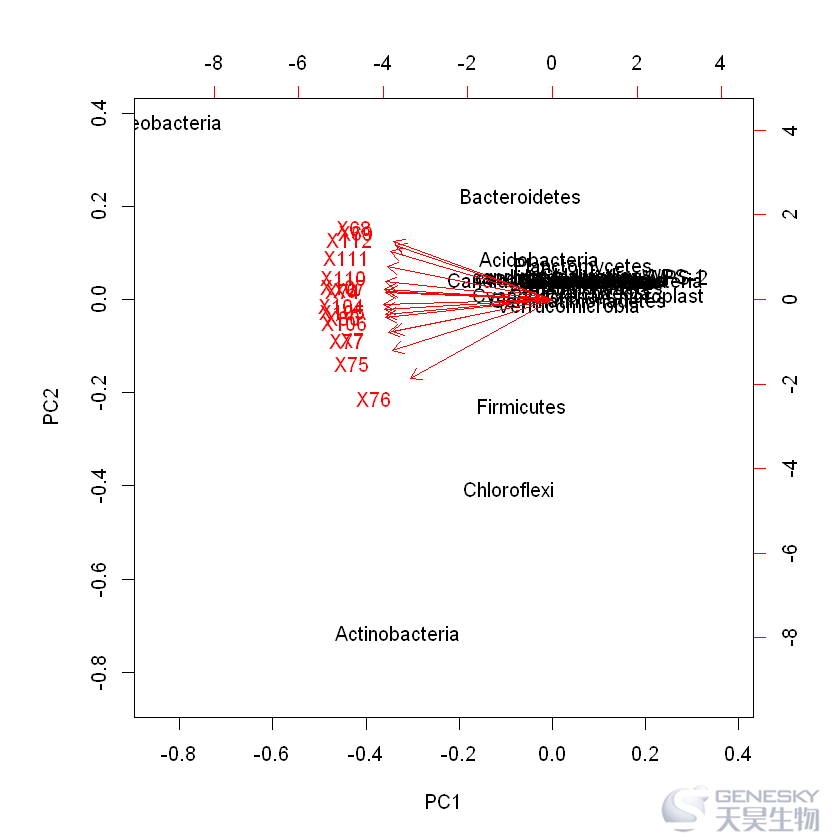
二
ggbiplot-PCA作图
install_github 需要安装软件rtools(不是R包),最新的是rtools40,针对的是R 4.0 版本,一般安装的R version 3.6.*,所以rtools要选择版本下载,默认安装到C盘,避免麻烦。
1.加载R包
In[5]:
library(devtools)
library(ggbiplot)
2.读取数据
In[6]:
df = read.table('ggbiplot_data.txt',header = T,sep = ' ',row.names = 1)
head(df)
Out[6]:
Veillonella Neisseria Prevotella Porphyromonas Streptococcus Lachnoanaerobaculum Treponema Leptotrichia
UC01 0.01339965 0.16395553 0.08344061 0.002750146 0.1247513 0.000000000 0.053481568 0.04324166
UC02 0.05143359 0.13288473 0.07963721 0.065652428 0.1256290 0.006260971 0.003861908 0.03282621
UC03 0.05418373 0.04967817 0.14101814 0.017846694 0.1716208 0.006085430 0.015740199 0.02387361
UC04 0.05149210 0.01141018 0.10854301 0.004505559 0.2196021 0.007021650 0.032358104 0.02527794
UC05 0.04031597 0.09520187 0.08759509 0.075775307 0.2674664 0.001696899 0.001930954 0.02375658
UC06 0.05839672 0.02293739 0.11854886 0.003393798 0.1412522 0.005032183 0.018607373 0.03089526
3.指定绘图中的标签排列
In [7]:
Group = read.table('ggboxplot_group.txt',header = T,sep = ' ')
group = factor(Group$group,levels = c("CD","UC","HC"))
group
Out[7]:
UC UC UC UC UC UC UC UC UC UC CD CD CD CD CD CD CD CD CD CD CD CD HC HC HC HC HC HC HC HC
4. PCA分析
In [8]:
df_pca = prcomp(df,scale. = T)
df_pca
Out[8]:
Standard deviations (1, .., p=8):
1.7821933 1.2878327 1.0918639 0.7871798 0.7236448 0.6625366 0.4725125
0.4093539
Rotation (n x k) = (8 x 8):
PC1 PC2 PC3 PC4 PC5
Veillonella 0.4573712 -0.24397676 0.18983692 -0.3538455 0.23524282
Neisseria -0.2236327 -0.39722188 -0.52758737 0.1988236 0.61197685
Prevotella 0.3992054 0.34811232 0.03861493 -0.2718197 0.61713984
Porphyromonas -0.3377305 -0.42471400 0.21734617 -0.1666548 0.12510516
Streptococcus -0.1964093 0.10167920 0.71839929 0.5098384 0.36876531
Lachnoanaerobaculum 0.4728501 -0.21213688 0.11285891 0.2122485 -0.16521656
Treponema -0.1520557 0.64532333 -0.24043181 0.1116204 0.09942415
Leptotrichia 0.4267181 -0.09665939 -0.22414483 0.6451039 -0.02151154
PC6 PC7 PC8
Veillonella -0.03135291 0.12042148 0.70655408
Neisseria 0.15586753 -0.25749323 0.09597301
Prevotella -0.07492942 0.06084414 -0.50388271
Porphyromonas -0.74952775 0.15434102 -0.17110997
Streptococcus 0.14946847 -0.05381796 0.11758812
Lachnoanaerobaculum -0.25907424 -0.74950230 -0.13211541
Treponema -0.51264597 -0.20198211 0.42033615
Leptotrichia -0.23462707 0.53500084 -0.02074083
5.绘图
In[9]:
ggbiplot(df_pca, obs.scale = 1, var.scale = 1, groups = group, ellipse = TRUE, circle = TRUE) + scale_color_discrete(name = '') + theme(legend.direction = 'horizontal', legend.position = 'top')
Out[9]:
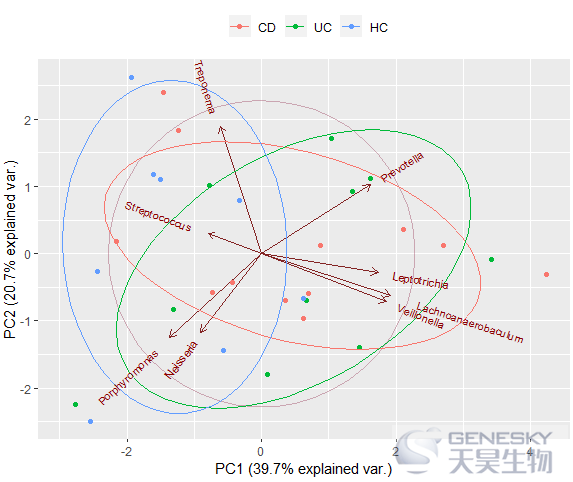
var.axes= T/F 是否添加物种及箭头
In[10]:
ggbiplot(df_pca, obs.scale = 1, var.scale = 1, groups = group, ellipse = TRUE,var.axes = F)
Out[10]:
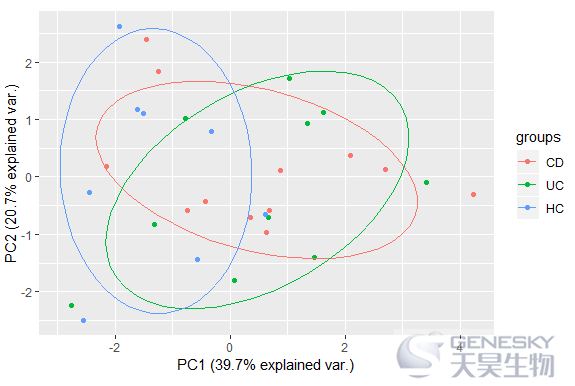
In [11]:
ggbiplot(df_pca, obs.scale = 1, var.scale = 1, groups = group, ellipse = TRUE,var.axes = T)
Out[11]:
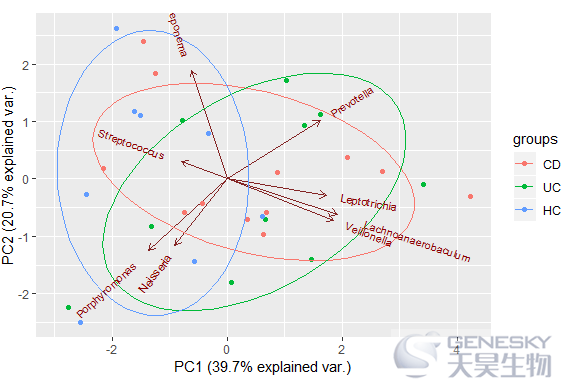
往期相关链接:
3分钟学会CHIP-seq类实验测序数据可视化 —IGV的使用手册;
10分钟搞定多样性数据提交,最快半天内获取登录号,史上最全的多样性原始数据提交教程;
【WGS服务升级】人工智能软件SpliceAI助力解读罕见和未确诊疾病中的非编码突变;
20分钟搞定GEO上传,史上最简单、最详细的GEO数据上传攻略;
【本群将为大家提供】
分享生信分析方案
提供数据素材及分析软件支持
定期开展生信分析线上讲座

QQ号:1040471849
作者:大熊
审核:有才
来源:天昊生信团

创新基因科技,成就科学梦想
微信扫一扫
关注该公众号
前往“发现”-“看一看”浏览“朋友在看”