
 咨询热线:400-065-6886
咨询热线:400-065-6886

 咨询热线:400-065-6886
咨询热线:400-065-6886
0一
数据预处理
In [1]:
df = read.table('taxon.lefse.result.xls',sep = ' ',header = T)
head(df,4)
Out[1]:
Taxonomy Group LDA Pvalue
k__Archaea A 6.879678 0.03899022
k__Archaea.p__Euryarchaeota B 5.548066 0.03899022
k__Archaea.p__Euryarchaeota.c__Thermoplasmata B 5.497864 0.03899022
k__Archaea.p__Euryarchaeota.c__Thermoplasmata.o__Methanomassiliicoccales B 5.514124 0.02732372
如上图所示,我们拿到的lefse的文件格式一般是这个样子的,为了减少文字长度,我们通常只显示最后一个分类层级的名称,需要用“.” 作为分隔符,取最后一列。linux上使用awk -F '.' '{print $NF}' taxon.lefse.result.xls 获取。结果如下所示:
df = read.table('lefse.result.xls',sep = ' ',header = T)head(df)Taxonomy Group LDA Pvaluek__Archaea A 6.879678 0.0389902
2p__Euryarchaeota B 5.548066 0.03899022c__Thermoplasmata B 5.497864 0.03899022o__Methanomassiliicoccales B 5.514124 0.02732372f__Methanomassiliicoccaceae B 5.501945 0.02732372g__Methanomassiliicoccus B 5.520169 0.02732372
二
Lefse绘图
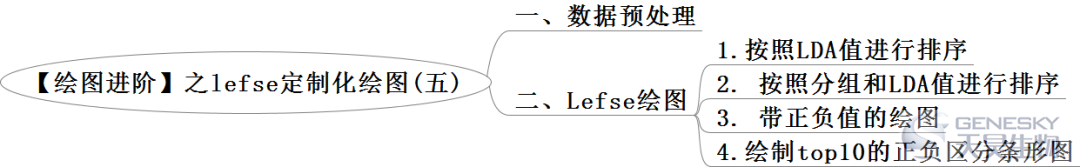
1.按照LDA值进行排序
In [4]:
ggplot(df,aes(x = reorder(Taxonomy,LDA),y=LDA,fill = Group)) +
geom_bar(stat = 'identity',colour = 'black',width = 0.9,position = position_dodge(0.7))+
xlab('') + ylab('LDA') + coord_flip() +
theme_bw() +
theme(panel.grid.major.y = element_blank()) + theme(axis.text.y = element_blank(),
axis.ticks = element_blank(), axis.text.x = element_text(face = 'bold'),axis.title.x = element_text(face='bold')) +
theme(panel.border = element_blank())+
geom_text(aes(y = ifelse(df$LDA >0,-0.1,0.1),label=Taxonomy),fontface=4,size=1,hjust = ifelse(df$LDA>0,1,0))
Out[4]:
Taxonomy Group LDA Pvalue161 g__Oceanibaculum A 3.942752 0.02413390147 s__Methylobacterium_oxalidis A 4.063120 0.0379415471 s__Actinocorallia_longicatena A 4.134189 0.04593813116 s__Paenibacillus_edaphicus A 4.138317 0.0389902226 s__Aquihabitans_daechungensis A 4.149702 0.0496478088 g__Segetibacter A 4.174600 0.03473526
In [6]:
df$Taxonomy = factor(df$Taxonomy,levels = as.character(df$Taxonomy))
In [7]:
df$Taxonomy = factor(df$Taxonomy,levels = as.character(df$Taxonomy))
ggplot(df,aes(x = Taxonomy,y=LDA,fill = Group)) +
geom_bar(stat = 'identity',colour = 'black',width = 0.9,position = position_dodge(0.7))+
xlab('') + ylab('LDA') + coord_flip() +
theme_bw() +
theme(panel.grid.major.y = element_blank()) +
theme(axis.text.y = element_blank(),axis.ticks = element_blank(),
axis.text.x = element_text(face = 'bold'),axis.title.x = element_text(face='bold')) +
theme(panel.border = element_blank())+
geom_text(aes(y = ifelse(df$LDA >0,-0.1,0.1),label=Taxonomy),fontface=4,size=1,hjust = ifelse(df$LDA>0,1,0))
Out[7]:

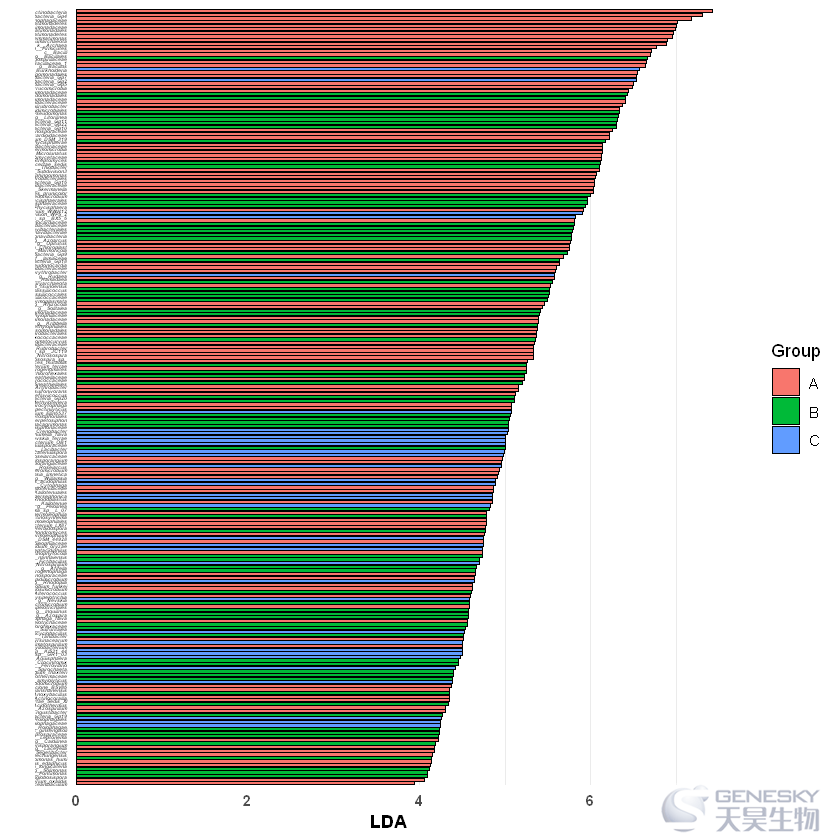
将B组的LDA值改成负数Taxonomy Group LDA Pvalue161 g__Oceanibaculum A -3.942752 0.02413390147 s__Methylobacterium_oxalidis A -4.063120 0.0379415471 s__Actinocorallia_longicatena A -4.134189 0.04593813116 s__Paenibacillus_edaphicus A -4.138317 0.0389902226 s__Aquihabitans_daechungensis A -4.149702 0.0496478088 g__Segetibacter A -4.174600 0.03473526ggplot(df,aes(x = Taxonomy,y=LDA,fill = Group)) + geom_bar(stat = 'identity',colour = 'black',width = 0.9,position = position_dodge(0.7))+ xlab('') + ylab('LDA') + coord_flip() + theme_bw() + theme(panel.grid.major.y = element_blank()) + theme(axis.text.y = element_blank(),axis.ticks = element_blank(), axis.text.x = element_text(face = 'bold'),axis.title.x = element_text(face='bold')) + theme(panel.border = element_blank()) + geom_text(aes(y = ifelse(df$LDA >0,-0.1,0.1),label=Taxonomy),fontface=4,size=1,hjust = ifelse(df$LDA>0,1,0))
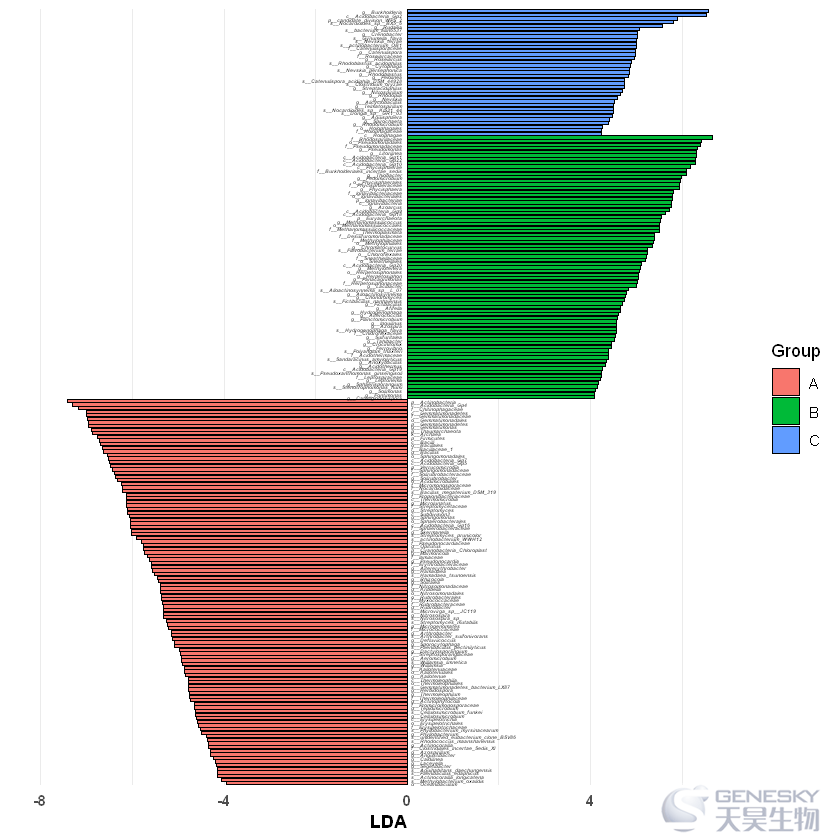
df = rbind(head(df[df$Group=='A',],10),head(df[df$Group=='B',],10),head(df[df$Group=='C',],10))head(df)Taxonomy Group LDA Pvalue161 g__Oceanibaculum A -3.942752 0.02413390147 s__Methylobacterium_oxalidis A -4.063120 0.0379415471 s__Actinocorallia_longicatena A -4.134189 0.04593813116 s__Paenibacillus_edaphicus A -4.138317 0.0389902226 s__Aquihabitans_daechungensis A -4.149702 0.0496478088 g__Segetibacter A -4.174600 0.03473526
通过geom_text(size=4) 调节字体大小
In [12]:
df$Taxonomy = factor(df$Taxonomy,levels = as.character(df$Taxonomy))
ggplot(df,aes(x = Taxonomy,y=LDA,fill = Group)) +
geom_bar(stat = 'identity',colour = 'black',width = 0.9,position = position_dodge(0.7))+
xlab('') + ylab('LDA') + coord_flip() +
theme_bw() +
theme(axis.text.y = element_blank(),axis.ticks = element_blank(),
axis.text.x = element_text(face = 'bold'),axis.title.x = element_text(face='bold')) +
theme(panel.border = element_blank()) +
geom_text(aes(y = ifelse(df$LDA >0,-0.1,0.1),label=Taxonomy),fontface=4,size=4,hjust = ifelse(df$LDA>0,1,0))
Out[12]:
往期相关链接:
1、R基础篇
2、R进阶
【绘图进阶】之六种带中心点的PCA 图和三维PCA图绘制(四);
【绘图进阶】之交互式可删减分组和显示样品名的PCA 图(三);
3、数据提交
3分钟学会CHIP-seq类实验测序数据可视化 —IGV的使用手册;
10分钟搞定多样性数据提交,最快半天内获取登录号,史上最全的多样性原始数据提交教程;
20分钟搞定GEO上传,史上最简单、最详细的GEO数据上传攻略;
4、表达谱分析
5、医学数据分析
【本群将为大家提供】
分享生信分析方案
提供数据素材及分析软件支持
定期开展生信分析线上讲座

QQ号:1040471849
作者:大熊
审核:有才
来源:天昊生信团
微信扫一扫
关注该公众号
前往“发现”-“看一看”浏览“朋友在看”